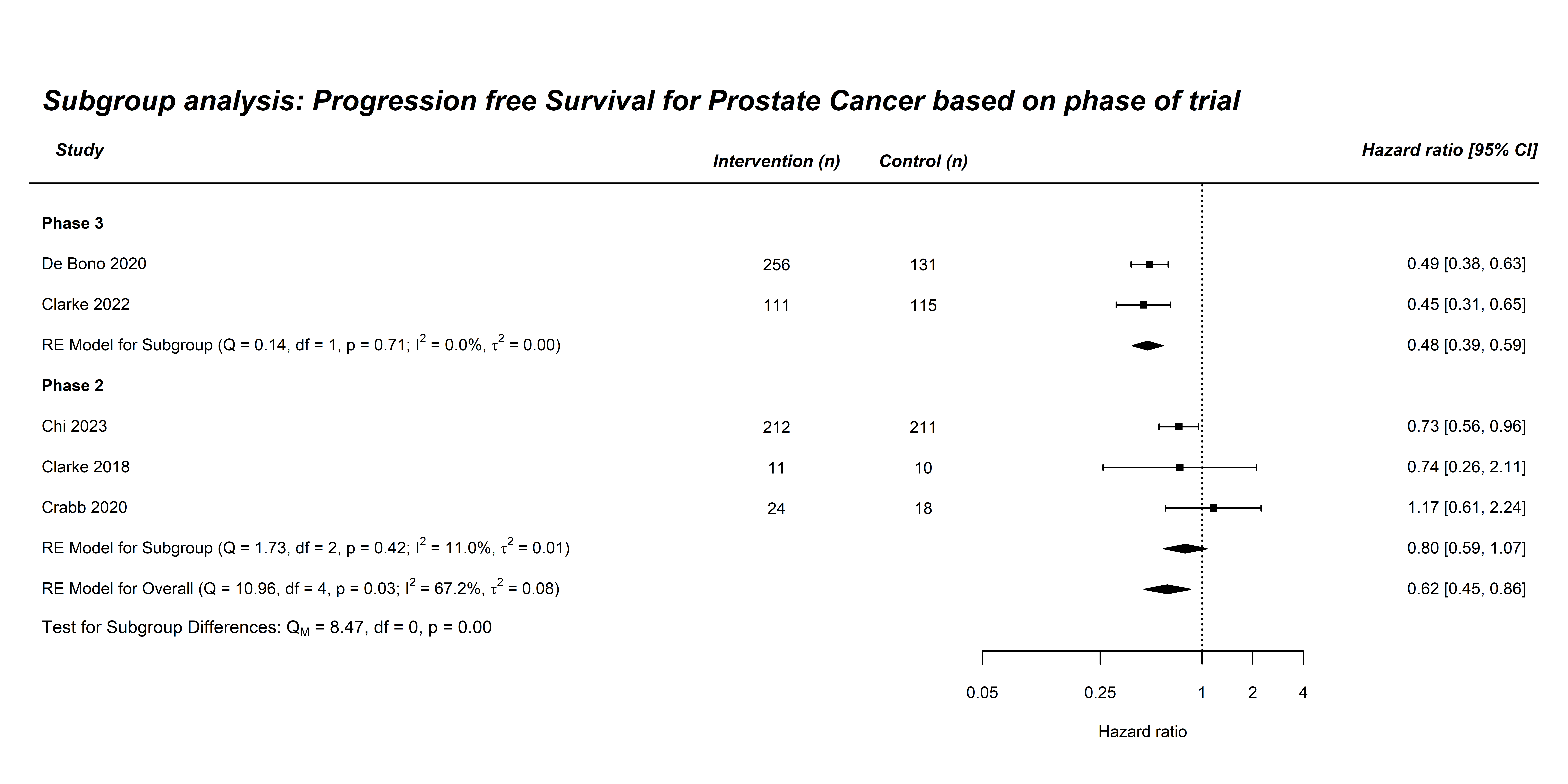Meta-analysis in R
Meta-analysis in R
Cord clamping in pre-term babies data from the recently published individual participant data meta analysis is used for this task.(Seidler et al. 2023)
Note
Note that only summary level data is used for this tutorial.
Code
Icomp <- read_excel("~/GitHub/Profile/Icomp.xlsx")Calculate log odd ratios and corresponding sampling variances
Random-effects model (using log odd ratios and variances as input)
Helper function
Code
mlabfun <- function(text, res) {
list(bquote(paste(.(text),
" (Q = ", .(formatC(res$QE, digits=2, format="f")),
", df = ", .(res$k - res$p),
", p ", .(metafor:::.pval(res$QEp, digits=2, showeq=TRUE, sep=" ")), "; ",
I^2, " = ", .(formatC(res$I2, digits=1, format="f")), "%, ",
tau^2, " = ", .(formatC(res$tau2, digits=2, format="f")), ")")))}Forest plot
Code
data_height <- nrow(dat)
#change the left bound after you have run the forest plot once
left_bound <- -8.5
#change the right bound after you have run the forest plot once
right_bound <-4
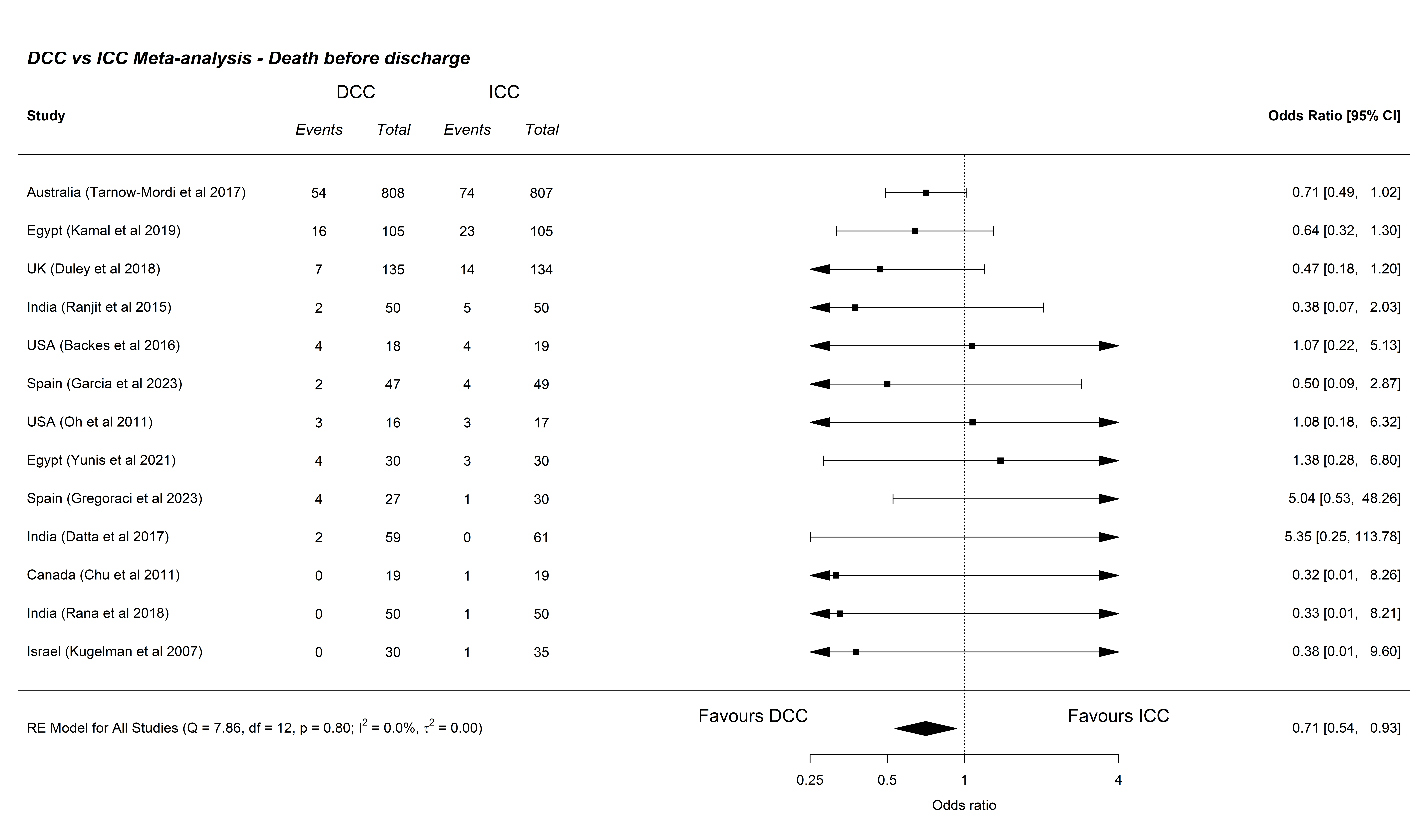
sav <- forest(res, header=TRUE, xlim=c(left_bound,right_bound), at=log(c(0.5, 0.25, 1, 4)), atransf=exp, ylim=c(-1, (data_height +3)), xlab ="Odds ratio", mlab=mlabfun("RE Model for All Studies", res), ilab=cbind(Icomp$`n_event DCC`, N_DCC, Icomp$`n_event ICC`, Icomp$N_ICC), ilab.xpos=seq(-5.8,-3.8, length = 4), slab=paste(dat$studyid), psize=1)
text(left_bound, data_height+3.5, pos=4, cex=1.3, c("DCC vs ICC Meta-analysis - Death before discharge"), font = 4)
text(sav$ilab.xpos, (data_height+2), pos=1, c("Events","Total","Events","Total"), cex = 1.1, font =3)
text(c(mean(sav$ilab.xpos[1:2]),mean(sav$ilab.xpos[3:4])), data_height+3, c("DCC","ICC"), pos=1, cex=1.3)
text(c(log(0.15),log(4)), -0.3, pos=1, c("Favours DCC", "Favours ICC"), cex=1.3)Sensitivity analysis: Leave one out meta-analysis
Code
data_height <- nrow(dat)
#change the left bound after you have run the forest plot once
left_bound <- -8.5
#change the right bound after you have run the forest plot once
right_bound <-4
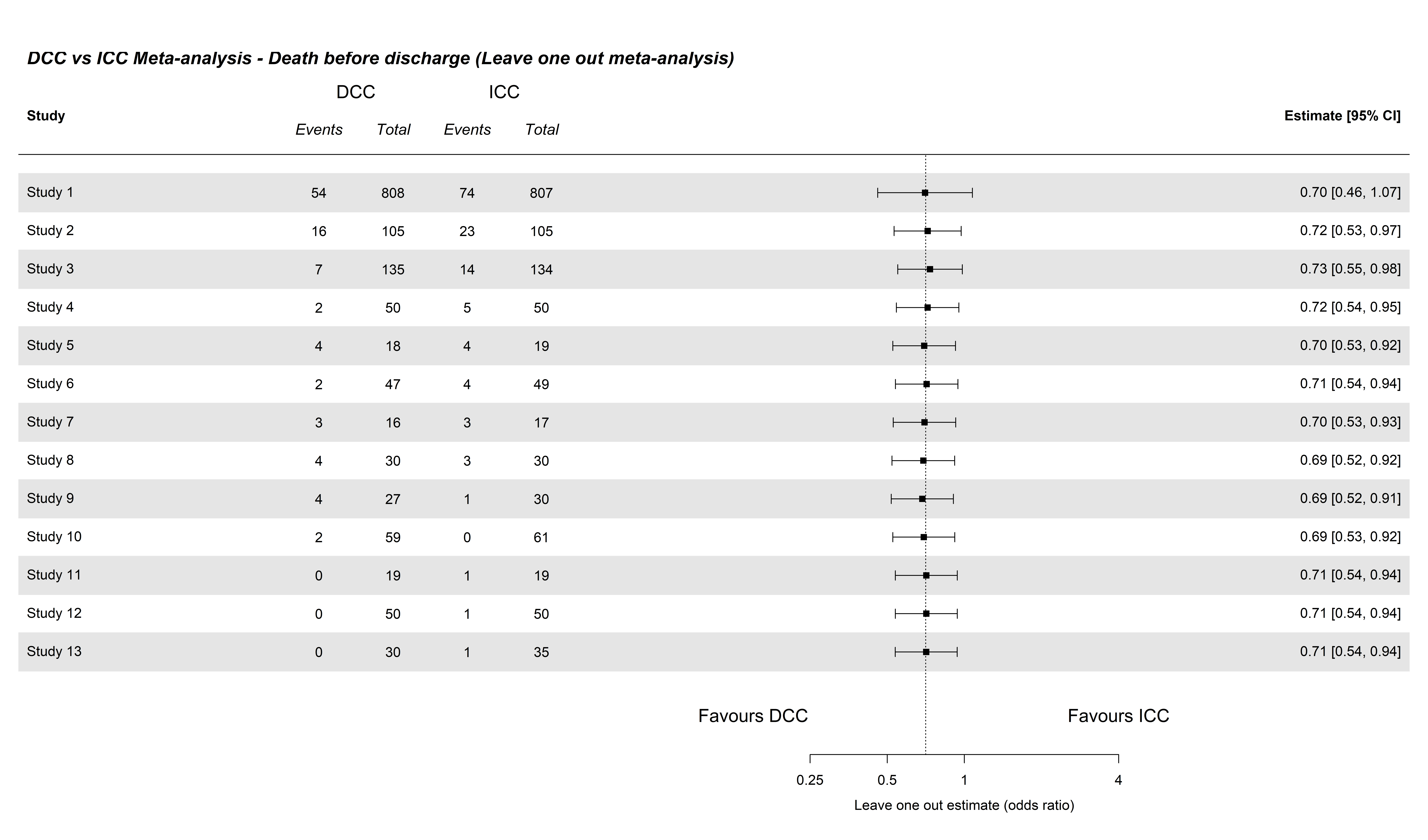
sav <- forest(res1$estimate, sei=res1$se, header=TRUE, xlim=c(left_bound,right_bound), at=log(c(0.5, 0.25, 1, 4)), shade=TRUE, atransf=exp, ylim=c(-1, (data_height +3)), xlab ="Leave one out estimate (odds ratio)", ilab=cbind(Icomp$`n_event DCC`, Icomp$N_DCC, Icomp$`n_event ICC`, Icomp$N_ICC), ilab.xpos=seq(-5.8,-3.8, length = 4), refline=coef(res), psize=1)
text(left_bound, data_height+3.5, pos=4, cex=1.3, c("DCC vs ICC Meta-analysis - Death before discharge (Leave one out meta-analysis)"), font = 4)
text(sav$ilab.xpos, (data_height+2), pos=1, c("Events","Total","Events","Total"), cex = 1.1, font =3)
text(c(mean(sav$ilab.xpos[1:2]),mean(sav$ilab.xpos[3:4])), data_height+3, c("DCC","ICC"), pos=1, cex=1.3)
text(c(log(0.15),log(4)), -0.3, pos=1, c("Favours DCC", "Favours ICC"), cex=1.3)Sensitivity analysis: Cumulative meta-analysis
Code
data_height <- nrow(dat)
#change the left bound after you have run the forest plot once
left_bound <- -8.5
#change the right bound after you have run the forest plot once
right_bound <-4
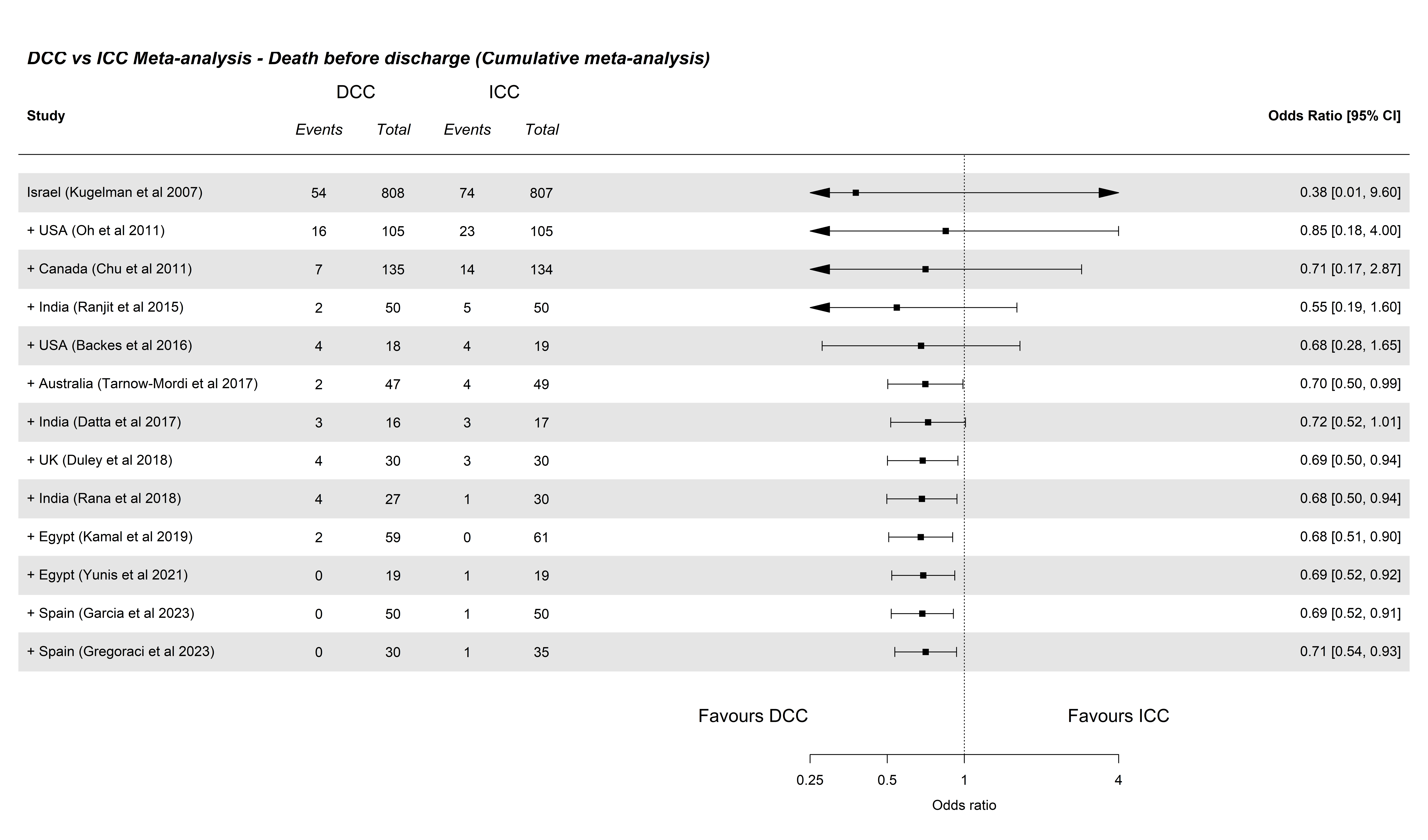
sav <- forest(res2, header=TRUE, xlim=c(left_bound,right_bound), at=log(c(0.5, 0.25, 1, 4)), shade=TRUE, atransf=exp, digits=c(2L,3L), ylim=c(-1, (data_height +3)), xlab ="Odds ratio", ilab=cbind(Icomp$`n_event DCC`, Icomp$N_DCC, Icomp$`n_event ICC`, Icomp$N_ICC), ilab.xpos=seq(-5.8,-3.8, length = 4), psize=1)
text(left_bound, data_height+3.5, pos=4, cex=1.3, c("DCC vs ICC Meta-analysis - Death before discharge (Cumulative meta-analysis)"), font = 4)
text(sav$ilab.xpos, (data_height+2), pos=1, c("Events","Total","Events","Total"), cex = 1.1, font =3)
text(c(mean(sav$ilab.xpos[1:2]),mean(sav$ilab.xpos[3:4])), data_height+3, c("DCC","ICC"), pos=1, cex=1.3)
text(c(log(0.15),log(4)), -0.3, pos=1, c("Favours DCC", "Favours ICC"), cex=1.3)Meta analysis with subgroups
Data from Cochrane review: Next-generation sequencing for guiding matched targeted therapies in people with relapsed or metastatic cancer(Kazmi et al. 2023) is used for this task.
Random-effects model (using log hazard ratios and variances as input)
Helper function
Code
mlabfun <- function(text, res4) {
list(bquote(paste(.(text),
" (Q = ", .(formatC(res4$QE, digits=2, format="f")),
", df = ", .(res4$k - res4$p),
", p ", .(metafor:::.pval(res4$QEp, digits=2, showeq=TRUE, sep=" ")), "; ",
I^2, " = ", .(formatC(res4$I2, digits=1, format="f")), "%, ",
tau^2, " = ", .(formatC(res4$tau2, digits=2, format="f")), ")")))}Forest plot
Code
sav <-forest(res, xlim=c(-16, 4.6), at=log(c(0.05, 0.25, 1,2,4)), atransf=exp,
cex=0.75, ylim=c(-1, 12), order=dat$alloc, rows=c(2:4, 7:8), xlab ="Hazard ratio",
addfit=FALSE, ilab=cbind(df$int, df$cont), ilab.xpos=seq(-5.8,-3.8, length = 2),
psize=1
)
### set font expansion factor (as in forest() above) and use a bold font
op <- par(cex=0.75, font=2)
### add text for the subgroups
text(-16, c(5, 9), pos=4, c("Phase 2", "Phase 3"))
### set par back to the original settings
par(op)
### fit random-effects model in the two subgroups and overll pooled result
res1 <- rma(yi, vi, subset=(phase=="3"), data=dat)
res2 <- rma(yi, vi, subset=(phase=="2"), data=dat)
res3 <- rma(yi, vi, data=dat)
### add summary polygons for the two subgroups and overall pooled result
addpoly(res3, row= 0, mlab=mlabfun("RE Model for Overall", res3))
addpoly(res2, row= 1, mlab=mlabfun("RE Model for Subgroup", res2))
addpoly(res1, row= 6, mlab=mlabfun("RE Model for Subgroup", res1))
text(-16, 12, pos=4, cex=1.3, c("Subgroup analysis: Progression free Survival for Prostate Cancer based on phase of trial"), font = 4)
text(2, 10.8, pos=4, cex=0.8, c("Hazard ratio [95% CI]"), font = 4)
text(sav$ilab.xpos, 11, pos=1, c("Intervention (n)","Control (n)"), cex = 0.8, font =4)
text(c(log(0.05),log(4)), -1.5, pos=1, c("Favours Intervention", "Favours Control"), cex=0.8)
text(-14.8, 10.8, "Study", cex=0.8, pos = 2, font =4)
### add text for the test of subgroup differences
text(-16, -1, pos=4, cex=0.8, bquote(paste("Test for Subgroup Differences: ",
Q[M], " = ", .(formatC(res$QM, digits=2, format="f")), ", df = ", .(res$p - 1),
", p = ", .(formatC(res$QMp, digits=2, format="f")))))Network Meta-analysis in R
we use the TherapyFormats data. This data set is modeled after a real network meta-analysis assessing the effectiveness of different delivery formats of cognitive behavioral therapy for depression (Cuijpers et al. 2019).
Code
install.packages ("netmeta")The following package(s) will be installed:
- netmeta [2.9-0]
These packages will be installed into "~/GitHub/Profile/renv/library/windows/R-4.4/x86_64-w64-mingw32".
# Installing packages --------------------------------------------------------
- Installing netmeta ... OK [linked from cache]
Successfully installed 1 package in 54 milliseconds.Code
library (netmeta)
if (!require("remotes")) {
install.packages("remotes")
}
remotes::install_github("MathiasHarrer/dmetar")Creating netwotrk meta analysis graph
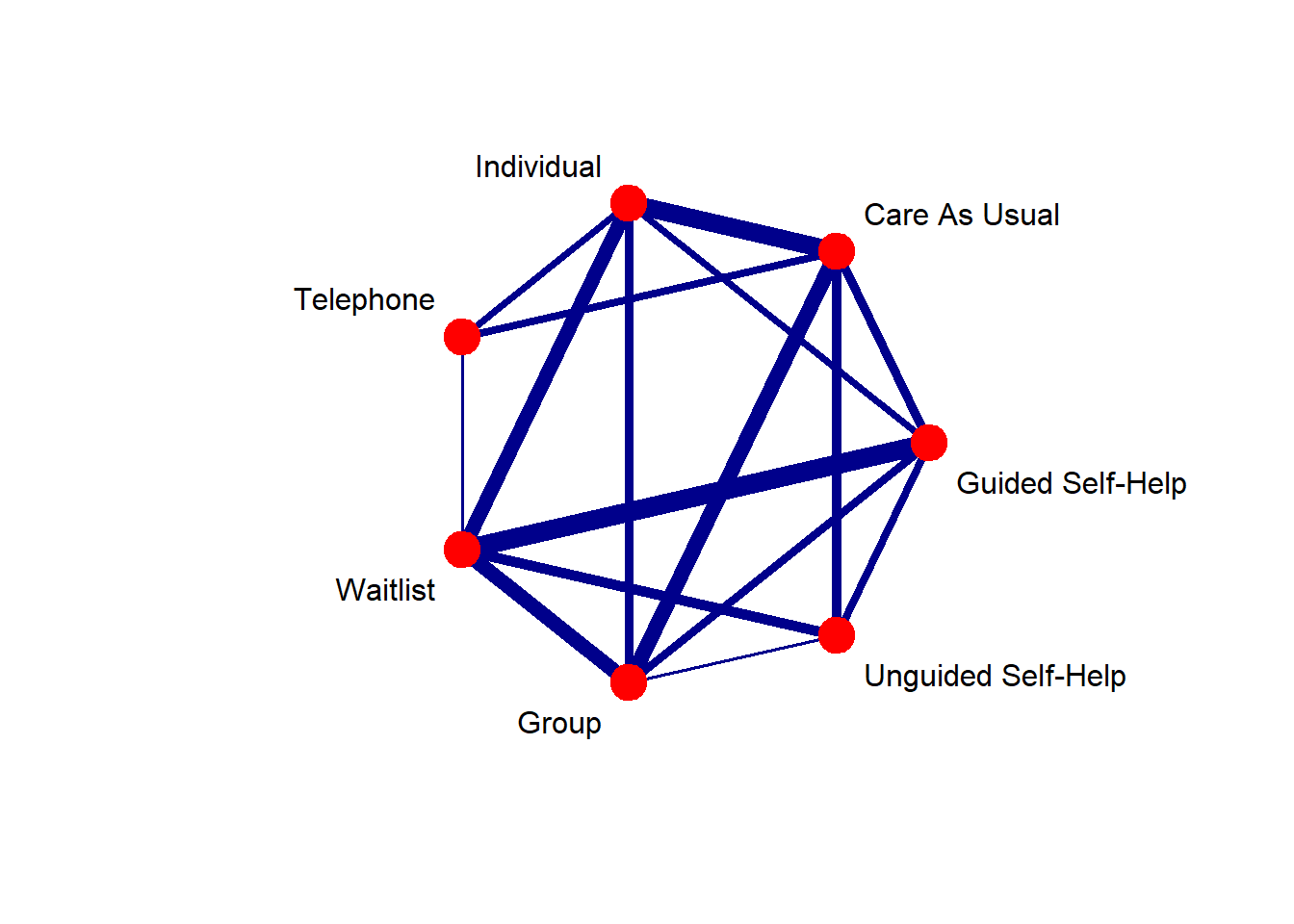
Code
net <- netmeta (TE, seTE, treat1, treat2, author, data = TherapyFormats, sm= "SMD", fixed= TRUE, random= FALSE, reference ="cau")
# Replace with full name (see treat1.long and treat2.long)
long.labels <- c("Care As Usual", "Group",
"Guided Self-Help",
"Individual", "Telephone",
"Unguided Self-Help",
"Waitlist")
netgraph (net, plastic =FALSE, points=TRUE, col= "darkblue", thickness = "number.of.studies", lwd= 2.7, cex.points = 4, offset=0.05, scale = 1.1, col.points = "red", seq = 1, labels = long.labels)Ordered network effects forest plot (Pscore)
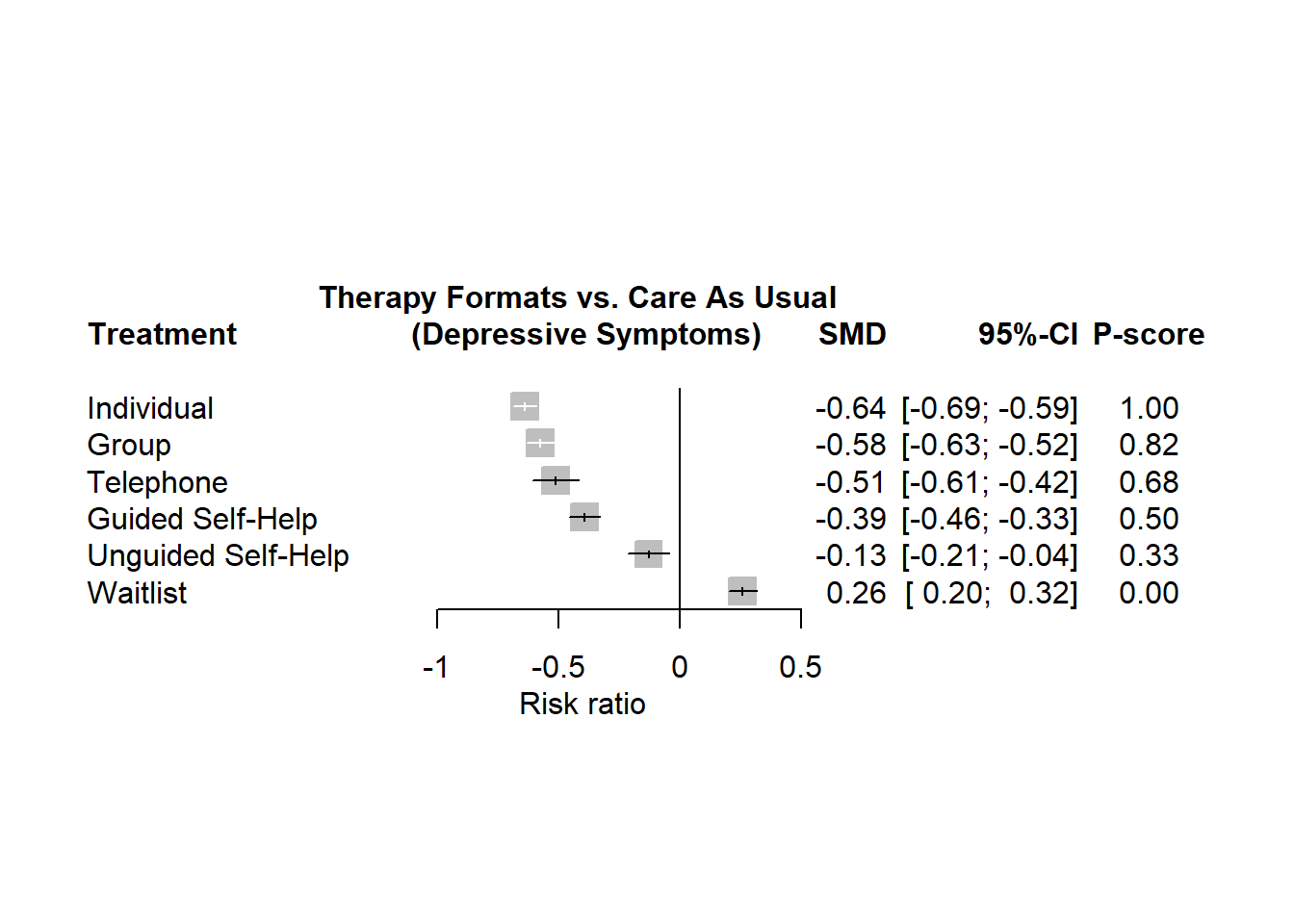
Code
forest(net, reference.group = "cau", sortvar = -Pscore, xlim = c(-1.3, 0.5),
smlab = paste("Therapy Formats vs. Care As Usual \n",
"(Depressive Symptoms)"), rightcols = c("effect", "ci", "Pscore"), xlab ="Risk ratio", direct= "TRURE", indirect ="FALSE", labels = long.labels, drop.reference.group = TRUE)Split the evidence into direct/indirect
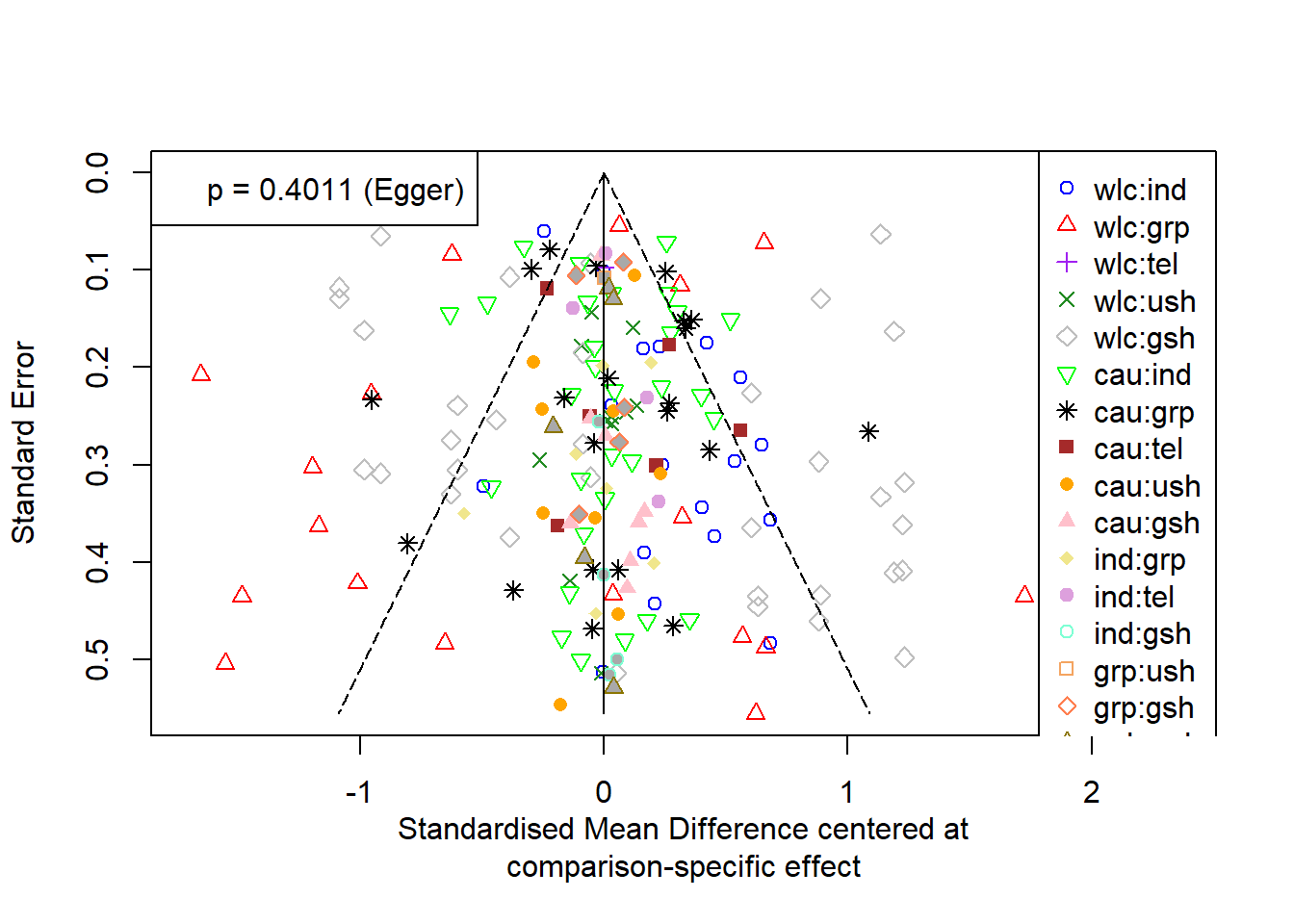
Publication bias
Code
funnel(net,
order = c("wlc", "cau", "ind", "grp", # from old to new
"tel", "ush", "gsh"),
pch = c(1:4, 5, 6, 8, 15:19, 21:24),
col = c("blue", "red", "purple", "forestgreen", "grey",
"green", "black", "brown", "orange", "pink",
"khaki", "plum", "aquamarine", "sandybrown",
"coral", "gold4"),
linreg = TRUE)References
Cuijpers, P., H. Noma, E. Karyotaki, A. Cipriani, and T. A. Furukawa. 2019. “Effectiveness and Acceptability of Cognitive Behavior Therapy Delivery Formats in Adults with Depression: A Network Meta-Analysis.” Journal Article. JAMA Psychiatry 76 (7): 700–707. https://doi.org/10.1001/jamapsychiatry.2019.0268.
Kazmi, F., N. Shrestha, T. Foord, T. F. Liu, P. Heesen, S. Booth, D. Dodwell, Y. Kheng Wei, S. Lord, and S. Blagden. 2023. “Are Targeted Therapies Effective in the Relapsed or Metastatic Cancer Setting? A Cochrane Meta-Analysis.” ESMO Open 8 (1): 101649. https://doi.org/10.1016/j.esmoop.2023.101649.
Seidler, Anna Lene, Mason Aberoumand, Kylie E Hunter, Angie Barba, Sol Libesman, Jonathan G Williams, Nipun Shrestha, et al. 2023. “Deferred Cord Clamping, Cord Milking, and Immediate Cord Clamping at Preterm Birth: A Systematic Review and Individual Participant Data Meta-Analysis.” The Lancet 402 (10418): 2209–22. https://doi.org/10.1016/s0140-6736(23)02468-6.